Faceted Search Tool for Pfizer CRF PDFs
A rules-based approach for extracting tabular information
I used a rules-based1 approach to extract tabular information from the Pfizer CRF PDF files2 and built a search tool.
Nearly this entire work is based on my online discussions with a_nineties reads so you need to read his article to understand why this tool is necessary and how it can help the people who are trying to analyze the Pfizer CRF files.
How to use the search tool
You will usually do all these steps to drill down into the information you need:
1 Download all the CSV files
These CSV files are organized by form type, as I mentioned in my previous article.
2 Get the required column names from the top level search tool
You can use this tool to filter by the fields which you see at the top of each page in a CRF file.
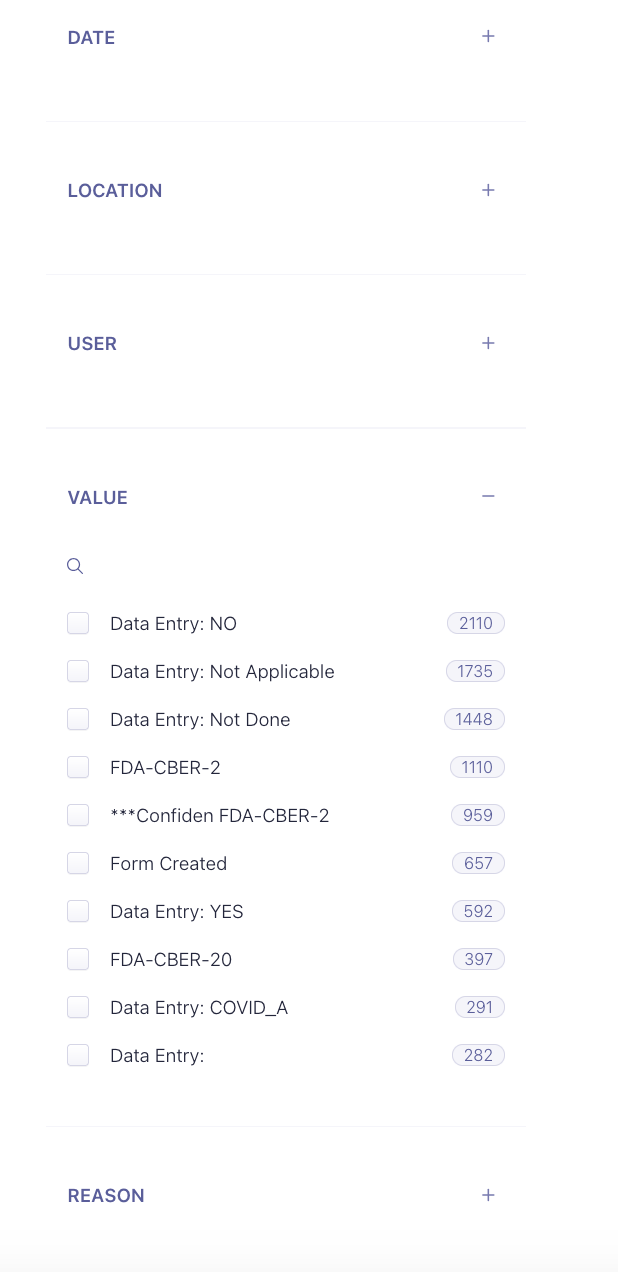
The top level search tool has a searchable filter called column names
These are the column names of the tables inside the PDF files. You will need to make note of the columns for a specific form type.
3 Use the columnar search tool to drill down into specific PDF files
A columnar search tool will consider only the columns of a specific type of table.
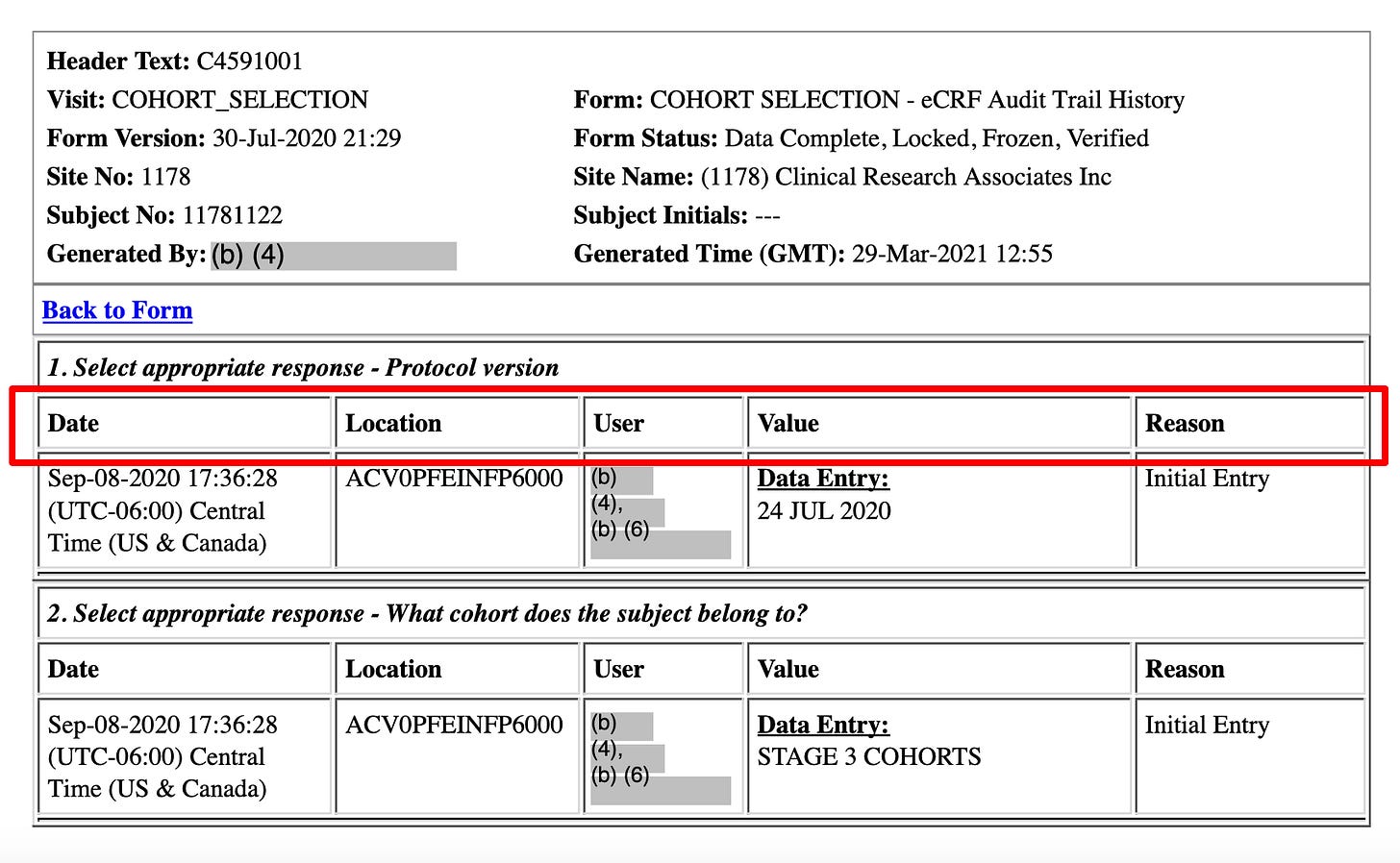
For example, this table type - with the column names Date, Location, User, Value and Reason - is very common in the CRF files
So I created a columnar search tool which has ONLY these columns as filters, in addition to the fields in the header.
As you can see, you can now search within these column values:
And you can jump directly to the corresponding PDF page on PHMPT by copy/pasting the URL (they are not clickable because I used Algolia’s default search interface instead of building my own)
4 Use the corresponding CSV file to complete your analysis
The first row in each search result is the name of the CSV file which has all the information in tabular format for the given form type.
You can open the corresponding CSV file and use it for more sophisticated analysis.
Here is an example CSV file which I loaded into Zoho Analytics
You can now filter down into whatever information you need within the CSV file.
For example, here is a comment from a_nineties on my previous article where he mentions that a form status which does not have the data “locked” is worthy of future investigation (emphasis mine)
already some interesting results in the csv, with a subset of pages missing the "data locked" designation in the header, for example subject 11171088. site 1117 was quite irregular in other regards, notably not entering adverse events deemed "reactogenic" into the AE log form. the CRFs are the most unadulterated form of data we have from the trial, which is why meticulous investigation of their contents is so important, and this endeavour is a colossal step in that direction. chapeau, eager to see further progress!
You can filter for this quite easily in the CSV files:
List of columnar search tools
I created columnar search tools for the top 3 frequently used column headers.
Search on Date, Location, User, Value Reason
Search on Item, Date, User, Comment
Search on Data Origin, Sample Type etc
Please let me know in the comments if you are interested in additional columnar search tools.
1 Please add a link to a sample page on PHMPT with the table. This will help me create suitable rules.
2 Please also add a small blurb as to why this could be useful. This can sometimes help brainstorm alternate ways to get the same result, or sometimes even write some code to automate the entire process.
Conclusion
I am well aware that the current data extraction process is quite suboptimal, especially with a lot of false positives for column name extraction and also a lot of poorly extracted table names.
But it still made sense to release this quickly to solicit more feedback instead of trying to overoptimize the data extraction. The process is very heavily reliant on rules at the moment, and it is pretty hard to come up with general rules which satisfy all the strange quirks in the layout of the CRF PDF files :-)
As opposed to a Machine Learning based approach. It is possible to use ML for this task and get better results, but for now I am just trying to build out the search tool as quickly as possible. I might do some optimizations in the future if people specifically request it.
At the moment, I have only included the Pfizer CRFs for 16+ years.











I just want to express my sincere thanks that you have used your gifts to help other researchers. It is generous and loving!
a crucial search term is the following (example: https://phmpt.org/wp-content/uploads/2023/08/125742_S1_M5_CRF_c4591001-1072-10721007.pdf, Page 64) form: FURTHER VACCINATION CONFIRMATION, with the text "eligible and NOT confirmed to have received only placebo at Vaccination 1/2". this is the trial arm assignment as recorded in the CRFs final submission state, which can differ from the assigned arm in the database (direct link to article section: https://modarnlife.substack.com/i/146474910/trial-arm-ambiguity). i found eight largely by chance, a systematic, software-based approch will surely find more!
another thing which might be interesting is checking for deleted adverse events not identifiable via AESPID gap (see @openvaet's article section here https://openvaet.substack.com/i/144275433/the-adverse-effects-to-covid-visit-re-qualification-was-widespread). essentially, the database's use of the CRF-specific adverse event identifier (1,2,3,4,5, etc) reveals events that were deleted, because the database wil only contain AESPIDs 1,4,5, just as an example. however, this exploit can't find deleted events if for example events 4 and 5 are deleted, so the maximum AESPID in the database is 3. a neat example of how silly this gets is 12601108, from my article: "The CRF only lists AESPID 1 and 5 with 2,3,4 deleted, while the ADAE entries include AESPID 6, 8, 9, 10, and 11; AESPID 7 was added and deleted after CRF data cut."
i can only echo carol's comment: thank you for your efforts!