Automating FOIA PDF Document Analysis
A quick brain dump to give you a preview of what is coming, and to get some feedback
I recently read this amazingly detailed article by a_nineties reads on the Pfizer CRF documents and (finally) realized what he was trying to do!1
So here is a quick brain dump of how this process can be automated, and I also invite some feedback from not only a_nineties but also others who may be interested in this topic.
Parsing tables in PDF files
It is now possible to use Python libraries to parse tabular information in PDF files.
I used a library called pdfplumber which is actually pretty good and does as well2 as any Document AI services provided by the giant tech companies like Google and Microsoft.
How I parsed the data
I extracted tabular information from each page of the CRF PDF file.
Every page has the following information on the top:
You can see that the top section has a lot of information - subject number, site number, visit type, form name etc.
I parse out this information by extracting them as lines of text - this is not only much more reliable than parsing out tabular information, it also provides a lot of summary-level information we need for further analysis (e.g. Subject ID).
I then organized the data by the Form type (I use a combination of form name plus visit name and convert it into a slug so that I can use it inside URLs in the future if I need to).
My original reason for categorizing by form type was to structure all the information into equal (column) sized tables for further analysis. But I realized that even within a specific form type, the number of columns is not consistent.
An explanation for the inconsistency could itself make for a very long post, and this is a good time to point out that automating the extraction of tabular data from PDF files is still quite a poorly solved problem. Results
There are over 1000 CRF files, but I only ran my script for 20 of them. It is somewhat time consuming to run the script and I would like to get some feedback from folks who are interested before I start doing it more thoroughly.
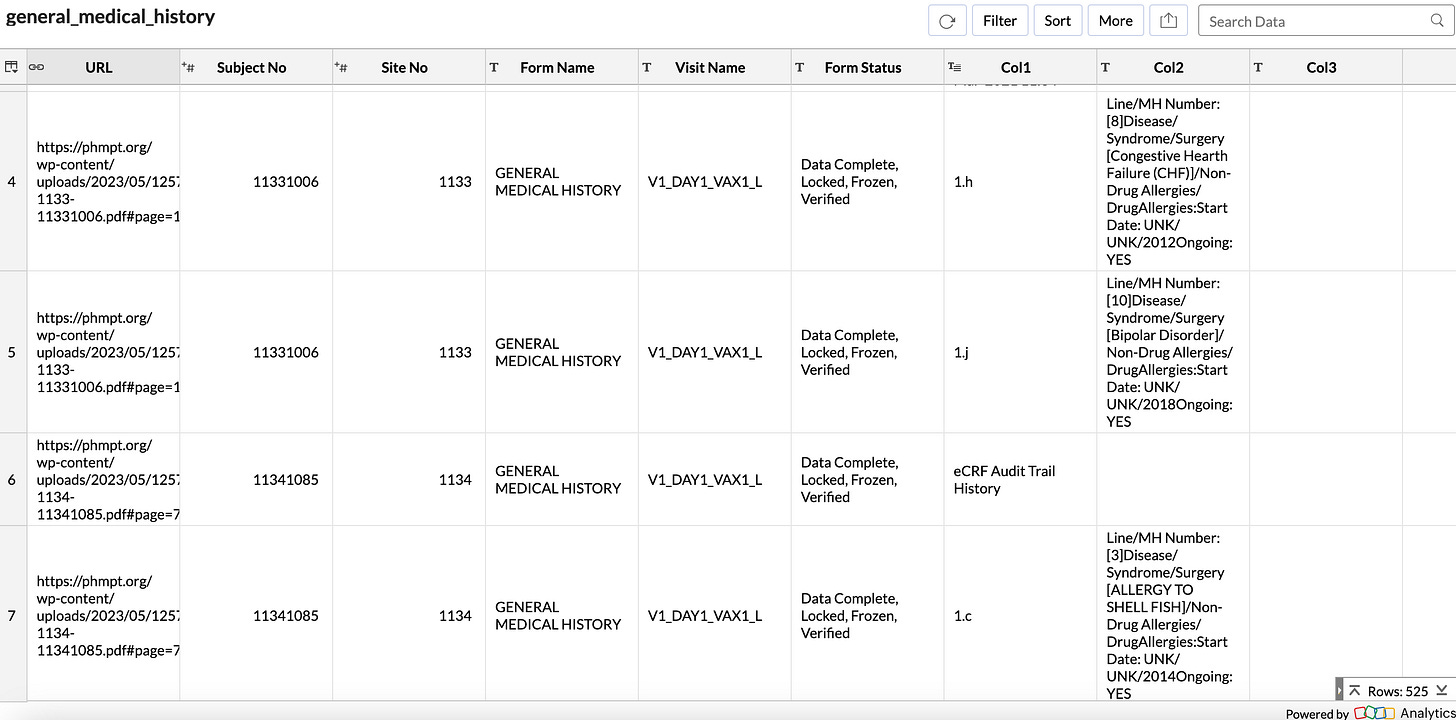
Here is an example CSV file for the Medical History Form Type
You can click on the URL and jump directly to the appropriate page on the PHMPT website.
I compiled all the parsed data into two types of CSV files: one based on subject number (subject ID), and the other based on form type.
How can we improve this process?
You might have noticed that sometimes the data is split in the wrong place. This is the big challenge with the existing Python libraries at the moment: we need to handle a lot of potential error cases because extracting tabular information from PDF files is still quite a hard problem.
I will keep making improvements to this process and keep updating these files over the next few weeks.
In the meantime, if you want me to parse some specific information, do let me know in the comments.
At least I think I fully understand it now. I am still not very sure.
One more casualty of the blanket refusal of the Data Science community to collaborate with people who do vaccine injury research is that we are only now getting a good picture of such requirements, more than 2 years after the first tranch of CRF FOIA files were published online in March 2022.
That is, the final parsed information provided by the paid services is not qualitatively better than the free Python library I use



PDFtoHTML.exe from xpdfreader gave the best results in terms of realistic PDF conversion, in my various attempts.
https://www.xpdfreader.com/download.html
If you have topics involving technical brainstorming, don't hesitate to have a_nineties organizing a call.
already some interesting results in the csv, with a subset of pages missing the "data locked" designation in the header, for example subject 11171088. site 1117 was quite irregular in other regards, notably not entering adverse events deemed "reactogenic" into the AE log form. the CRFs are the most unadulterated form of data we have from the trial, which is why meticulous investigation of their contents is so important, and this endeavour is a colossal step in that direction. chapeau, eager to see further progress!