Summary:
Foreign VAERS death reports often provide a summary for the “reported cause(s) of death”. This is much less common in US VAERS death reports.
We can write Python scripts to do string comparison and classify each reported cause to a symptom which has been recorded for that report in the SYMPTOMS CSV file
We can also use a Large Language Model (LLM) like the PaLM 2 API from Google to do this classification if simple string comparison is not sufficient
When you combine string comparison and LLM-based classification, it is possible to classify a large percent of the causes
Analysis of these causes shows that the number of deaths attributed to COVID19 itself (breakthrough COVID19) in foreign VAERS reports is less than 10% of all deaths
Recently I was trying to find out if VAERS reports mention the actual cause of death.
My research was prompted by this tweet.
I will come back to this tweet in the next part.
(I expect this to be a two part series where I continue to explain how we can solve this specific problem, and I hope I can also add some useful commentary about the benefits of using Large Language Models (LLMs) for VAERS analysis and research)
But first, let me clarify that this is about identifying the condition which caused the death.
This analysis is not about whether the vaccine caused the condition which led to the death.
While foreign VAERS reports rarely link the vaccine to the death, they are usually much more diligent about simply reporting the cause(s) of the death.
However, there isn’t any field in the VAERS CSV file which specifically lists the cause(s) of death from the full list of reported symptoms.
Here is an example VAERS report which has only two causes of death, but 15 recorded symptoms:
Symptoms: Arthralgia, Blood creatinine, Blood lactate dehydrogenase, Blood potassium, Blood urea, Body temperature, C-reactive protein, Cerebral haemorrhage, Haemoglobin, Neck pain, Platelet count, Prothrombin time, White blood cell count, Blood test, Blood pressure measurement
…Reported Cause(s) of Death: Cerebral haemorrhage; Pain from shoulder to neck; Pain from shoulder to neck; …
My goal is to
a) extract only the reported causes of death
b) where possible, map it to one of the coded symptoms
As you will see, creating such a dataset can also be immensely helpful for future text analysis of symptoms in VAERS writeups.
How often does foreign VAERS report the cause of death?
I did a simple search for the phrase “reported cause” in foreign vs US VAERS reports.
Here is the query for foreign VAERS reports
select count(*) as numdeaths,
count(*) filter ( where lower(symptom_text) like '%reported cause%' ) as numdeaths_with_reported_cause,
count(*) filter ( where lower(symptom_text) like '%caus%' ) as numdeaths_with_cause
from "NonDomesticVAERSDATA"
where died = 'Y'
and vaers_id in (select vaers_id
from "NonDomesticVAERSVAX"
where vax_type like 'COVID19%')And these are the results:
Here is the same query for US deaths
select count(*) as numdeaths,
count(*) filter ( where lower(symptom_text) like '%reported cause%' ) as numdeaths_with_reported_cause,
count(*) filter ( where lower(symptom_text) like '%caus%') as numdeaths_with_cause
from combined_vaersdata
where died = 'Y'
and vaers_id in (select vaers_id
from combined_vaersvax
where vax_type like 'COVID19%') And here are the results
As you can see, foreign VAERS reports are much more diligent about mentioning the cause of death.
With the stringent phrase match (‘reported cause’) we get 97.3% of deaths for foreign VAERS reports, and the more flexible match for the word ‘caus’ (which will catch multiple words like caused, causal, causation but will probably also have a lot of false positives) we get 99.4% of deaths.
For the US reports the corresponding numbers are just 12.8% and 21.8% respectively.
I will be using the foreign dataset from Nov 2022 for the rest of this analysis.
Comparing symptoms and cause of death
I first look at the sentence which contains the phrase “reported cause”.
Sometimes this will include multiple causes:
Reported Cause(s) of Death: could not walk even with assistance; high heart rate, particularly when standing; extremely out of breath (fighting for breath); off label use; immunisation; palpitations; pale; Severe shortness of breath; chest pain; interchange of vaccine products
So the script will parse each of these causes.
Notice that the same report has the following symptoms:
Chest pain, Computerised tomogram, Dyspnoea, Electrocardiogram, Gait inability, Heart rate increased, Immunisation, Pallor, Palpitations, X-ray, Off label use, Interchange of vaccine products, SARS-CoV-2 test
So the script will try to match each Reported Cause of Death to one of the symptoms which have been recorded for that particular VAERS report.
If this is not possible, then it will match it to “Unknown”.
Mapping reported cause to coded symptoms
One of the interesting Machine Learning problems in VAERS is being able to extract a lookup table to group similarly named symptoms.
That is a much broader problem, and is out-of-scope for this particular article.
But I mention it here, because there were so many different diseases caused or reactivated by the COVID19 vaccines, that it is actually possible to use the VAERS text writeups to create at least a condensed version of this lookup table.
And this is because there is something unique about VAERS death reports - because the problem is serious, both the “reported cause of death” sentence and the list of symptoms are usually very descriptive.
In turn, this means we can map the reported causes of death to the reported list of symptoms1 as long as we have some reasonable way to do it.
The first step is to obviously do a simple string comparison to see if you can find matches for all the reported causes of death.
But this is not always possible, because the writeup is usually just plain English.
For example, the writeup might say “could not walk even with assistance” and it is then “coded” in the SYMPTOMS CSV file as “Gait inability”.
So how do we map these plain English descriptions to the symptoms recorded for the given VAERS report?
Using a Large Language Model (LLM) to help with the mapping
Turns out, we can do it using PaLM2 (a competitor of ChatGPT)
Here is an example query sent to the PaLM 2 online demo (note: you need to be logged in to your Google account, and also create a Google cloud project, to be able to use the online demo. So I cannot provide a direct link to the tool)
Classify each reported cause into one of the following symptoms:
Chest pain, Computerised tomogram, Dyspnoea, Electrocardiogram, Gait inability, Heart rate increased, Immunisation, Pallor, Palpitations, X-ray, Off label use, Interchange of vaccine products, SARS-CoV-2 test
If the text doesn't fit any categories, classify it as the following:
Unknown
Classify each cause to a symptom in JSON format with the following fields:
cause, symptom
Reported causes:
could not walk even with assistance; high heart rate, particularly when standing; extremely out of breath (fighting for breath); off label use; immunisation; palpitations; pale; Severe shortness of breath; chest pain; interchange of vaccine products
Categories:And here is the response:
[
{
"cause": "could not walk even with assistance",
"symptom": "Gait inability"
},
{
"cause": "high heart rate, particularly when standing",
"symptom": "Heart rate increased"
},
{
"cause": "extremely out of breath (fighting for breath)",
"symptom": "Dyspnoea"
},
{
"cause": "off label use",
"symptom": "Off label use"
},
{
"cause": "immunisation",
"symptom": "Immunisation"
},
{
"cause": "palpitations",
"symptom": "Palpitations"
},
{
"cause": "pale",
"symptom": "Pallor"
},
{
"cause": "Severe shortness of breath",
"symptom": "Dyspnoea"
},
{
"cause": "chest pain",
"symptom": "Chest pain"
},
{
"cause": "interchange of vaccine products",
"symptom": "Interchange of vaccine products"
}
]As you can see, it can do a classification even for words which don’t appear in the symptom’s text. For example “could not walk even with assistance“ gets correctly mapped to the symptom “Gait inability”.
If there is no symptom matching the reported cause of death, PaLM can also map it to “Unknown”2
Here is an example (MESENTERIC VEIN TH gets classified as ‘Unknown’):
Classify each reported cause into one of the following symptoms:
Acute kidney injury, Cerebral haematoma, Cerebral haemorrhage, Coagulopathy, Computerised tomogram, Computerised tomogram head, Death, Hepatic failure, Hypovolaemic shock, Immunology test, Mesenteric vein thrombosis, Multiple organ dysfunction syndrome, Platelet count, Platelet factor 4, Pneumothorax, Portal vein thrombosis, Pulmonary embolism, Renal infarct, Renal vein thrombosis, Shock, Splenic infarction, Splenic vein thrombosis, Thrombocytopenia, Ultrasound scan, Venous occlusion
If the text doesn't fit any categories, classify it as the following:
Unknown
Classify each cause to a symptom in JSON format with the following fields:
cause, symptom
Reported causes:
HYPOVOLEMIC SHOCK; HEPATIC FAILURE; DEATH; RENAL INFARCTION; CEREBRAL HEMATOMA; PORTAL VEIN THROMBOSIS; VENOUS OBSTRUCTION; PULMONARY EMBOLISM; SPLENIC VEIN THROMBOSIS; CEREBRAL HEMORRHAGE; SPLENIC INFARCTION; MULTI ORGAN FAILURE; MESENTERIC VEIN TH
Categories:And here is the response from PaLM2:
[
{
"cause": "HYPOVOLEMIC SHOCK",
"symptom": "Shock"
},
{
"cause": "HEPATIC FAILURE",
"symptom": "Hepatic failure"
},
{
"cause": "DEATH",
"symptom": "Death"
},
{
"cause": "RENAL INFARCTION",
"symptom": "Renal infarct"
},
{
"cause": "CEREBRAL HEMATOMA",
"symptom": "Cerebral haematoma"
},
{
"cause": "PORTAL VEIN THROMBOSIS",
"symptom": "Portal vein thrombosis"
},
{
"cause": "VENOUS OBSTRUCTION",
"symptom": "Venous occlusion"
},
{
"cause": "PULMONARY EMBOLISM",
"symptom": "Pulmonary embolism"
},
{
"cause": "SPLENIC VEIN THROMBOSIS",
"symptom": "Splenic vein thrombosis"
},
{
"cause": "CEREBRAL HEMORRHAGE",
"symptom": "Cerebral haemorrhage"
},
{
"cause": "SPLENIC INFARCTION",
"symptom": "Splenic infarction"
},
{
"cause": "MULTI ORGAN FAILURE",
"symptom": "Multiple organ dysfunction syndrome"
},
{
"cause": "MESENTERIC VEIN TH",
"symptom": "Unknown"
}
]ChatGPT API vs PaLM 2
While you can also use the ChatGPT API (which is now at version GPT4) for this purpose, I chose PaLM2 for the following reasons:
a) The quality of the response is as good as GPT4 for this task
b) the output is more predictable and does not add unnecessary extra words. This is actually an important feature, since it makes it easier to programmatically parse the output
c) it is also cheaper than GPT-4 as of this writing3. It cost me less than $10 for processing the 15K reports for this task4.
There are also a couple of downsides to using PaLM-2 compared to GPT-4:
a) It is a little bit more tedious to set everything up for PaLM2- that is, GPT4 is much easier to go from zero to having some results
b) it is definitely oriented towards programmers at the moment, although I expect Google will soon provide tools to allow non-programmers to do similar analysis
Automating the process using the PaLM2 API
While you can use the PaLM demo tool and get this response for a single VAERS report, it does not make sense to manually do this for thousands of reports.
Since Google provides an API, you can write some Python code and automate the whole process.
Instead of sending a request to the PaLM2 API for every single report, I only ask for the classification when I cannot use simple string text matching to map each reported cause of death to one of the symptoms.
This way, I can save time and money on needless requests to the API, and also get a good understanding of how many reports permit this mapping using simple string comparison techniques5.
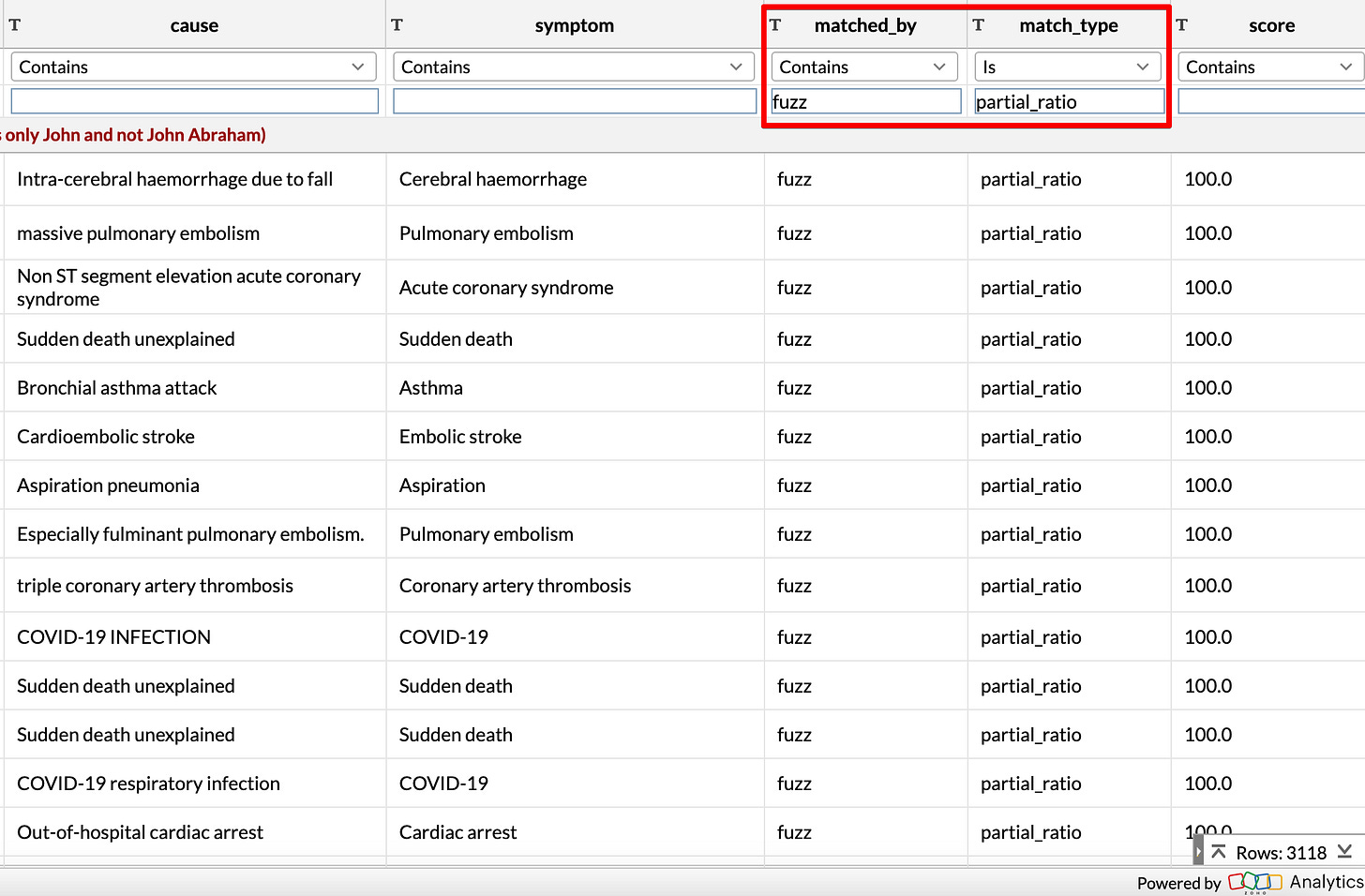
Here is the list of mappings produced by my Python code6:
String similarity comparison
First I try to compare the list of causes with the list of symptoms using simple string matching.
I used a Python library called FuzzyWuzzy (fuzzy means approximate in the context of string comparison) which has different ways of doing this matching:
Exact match
This is the case where the two words are almost exactly the same
Partial match
This is the case where the cause is a (almost) part of the symptom, or vice versa
Token sort match
This is a very interesting idea - where the algorithm first sorts all the letters in the cause and the symptom and compares them.
This can sometimes catch the stuff that is missed by both the ratio and the partial ratio algorithms. As you can see, this is especially useful for catching reordered phrases which are very similar.
This is the relevant code for the string comparison:
for cause in reported_causes:
lcause = cause.lower().strip()
if 'unknown' in lcause:
mapped_obj_list.append(
{
"cause": cause,
"symptom": 'Unknown'
}
)
has_match = True
matched_by = 'unknown'
break
has_match = False
for symptom in all_symptoms:
lsymptom = str(symptom).lower().strip()
score = fuzz.ratio(lcause, lsymptom)
if (score == 100) or (score >= 90 and len(lcause) >= 10):
print(f'Matched {cause} and {symptom}')
mapped_obj_list.append(
{
"cause": cause,
"symptom": symptom,
"match_type": "ratio",
"score": score
}
)
has_match = True
matched_by = 'fuzz'
break
score = fuzz.partial_ratio(lcause, lsymptom)
if (score == 100) or (score >= 95 and len(cause) >= 10):
print(f'Matched {cause} and {symptom}')
mapped_obj_list.append(
{
"cause": cause,
"symptom": symptom,
"match_type": "partial_ratio",
"score": score
}
)
has_match = True
matched_by = 'fuzz'
break
score = fuzz.token_sort_ratio(lcause, lsymptom)
if score >= 90:
print(f'Matched {cause} and {symptom}')
mapped_obj_list.append(
{
"cause": cause,
"symptom": symptom,
"match_type": "token_sort_ratio",
"score": score
}
)
has_match = True
matched_by = 'fuzz'
breakAs you can see, after checking for “Unknown”, the code moves from tightest (most stringent) match to the loosest (less stringent) match.
Using an LLM to help with the classification
If I am not able to map all the causes to a given symptom associated with the VAERS reports, I use an LLM to automate the task of doing this classification.
As it turns out, the particular task we are interested in - classifying a given list of causes to an already known list of symptoms - is something which is fairly easy for a large language model.
So I used Google’s PaLM API to do this classification, and you can see these mappings if you filter the dataset for matched_by = llm
A good way to see the power of using an LLM is to filter for a given symptom, and then filter for the “cause” field which does not contain that specific word
For e.g. here is what it looks like for Malaise
As you can see, this is the kind of stuff which is very tedious if you do it manually, but also can’t be mapped easily using the string comparison techniques as they use entirely different words.
Top symptoms associated with foreign death reports
So I created a list of mappings between the causes listed in the VAERS writeup and the symptoms associated with that particular death report7.
You can use these to identify the top symptoms associated with foreign VAERS deaths by creating a summary table.
As you can see, COVID-19 is reported as a cause of death in only 1128 of the 15K reports I have processed.
You also have a few more due to COVID-19 pneumonia, but the total foreign VAERS deaths which were directly attributed to COVID-19 and its after-effects is still less than 10% of the total number of reported deaths.
Conclusion
Based on the foreign VAERS death reports, you can already see that it is very unlikely that over 60% of the US VAERS deaths were due to COVID19.
This means if you really want to see how many of them were reported as being due to COVID198, you need to do more systematic analysis of the VAERS writeups instead of making blanket statements.
I will be covering this topic in more detail in the second part of this series.
But remember that it will not be a 1-to-1 mapping. Quite often, you will see VAERS reports with just one or two reported causes of death and well over 10 different symptoms.
Because LLMs are not 100% accurate, sometimes it does a wrong classification at this step. That is, it maps the reported cause to a wrong symptom when the correct classification should be “Unknown”
ChatGPT pricing is very dynamic and changes often, so it is possible this won’t be true when you read it
Note: this is because I parse the VAERS report using the spaCy NLP library and send only individual sentences, which reduces the size of the prompt quite a bit. This brings up an interesting question of why preprocessing input is a very important aspect of “prompt engineering”, and why it is a bad idea to use LLMs without even understanding the task. The creators of spaCy call this LLM Maximalism - the tendency to waste resources by turning everything into a giant prompt without thinking through your actual requirements
After running the code, I saw that about 50% of the reports can be resolved using simple string comparison
I could not process all the 16.5K foreign VAERS death reports since there were some API calls which returned some errors (due to some rate limite issues). I was able to process about 15K reports, so I could do the mapping for about 90% of the foreign VAERS death reports.
As I mentioned before, I wasn’t able to process my full dataset because I ran into some rate limits for the PaLM2 API, but I was able to process about 90% of them.
I am aware that some people ask if VAERS deaths which were attributed to COVID19 were simply vaccine induced deaths, but I am simply pointing out that even if you take the VAERS report at its word, the percentage calculation is probably wrong.