Anyone can fill out a VAERS report
But does that mean we should not be concerned about the sudden increase post COVID vaccination? What we can infer by looking at the amount of medical jargon in the text
We are often told that VAERS is unreliable because “anyone” can file a VAERS report.
But if you actually take a closer look, you can see that most of the reports are not just genuine, but there is enough medical jargon that it is almost impossible it was a layperson or an “anti-vaxxer” who did it just to increase the count in the VAERS database.
While the story of the “Incredible Hulk” report makes a good point, you only need to sift through a few reports to see that a vast majority of them don’t fit that pattern.
So is there any way to “quantify” the amount of medical jargon in a report?
To be clear, I am simply providing a method of inferring if the report was intended to be a prank or if it is authentic. Just because a report is authentic does not mean it establishes causation.
We will use a library called scispacy which has been trained on a vast corpus of medical literature and is in fact frequently used in BioNLP.
scispaCy is a Python package containing spaCy models for processing biomedical, scientific or clinical text.
I am looking for unique entities detected by the model called en_ner_bc5cdr_md because it can identify DISEASE and CHEMICAL entities. There are many more things you could identify by training your own model, but that is not necessary for the purpose of this article.
Identifying just these two types of entities gives us plenty of useful insight.
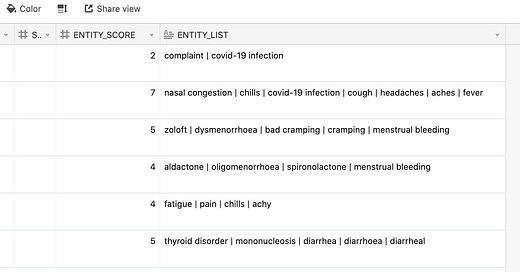
For each SYMPTOM_TEXT entry, I assign two values (that is, I add two additional columns into the dataframe)
ENTITY_LIST is the pipe-separated text value of all the unique entities identified by the model.
ENTITY_SCORE is the number of items in the entity list.
You can see the Python script here.
Here is an example
The higher the ENTITY_SCORE, the less likely that it is an inauthentic or a fake report.
And you can usually also glance at the report - remember that the first column is the URL field and clicking on it will take you directly to the snapshot of that VAERS report on the medalerts website - and check for yourself that this is the case. That is, a higher score for ENTITY_SCORE seems to indicate a more authentic report.
You can download the CSV files here.
How to verify
You can download the CSV files and check for yourself.
I have also added a Zoho Analytics Query Table view which allows you to verify this data online without having to download anything. It is also interactive, meaning you can do filter/sort/reorder etc. on the columns.
It consists of the reports from Jan, Feb and March 2022 where the ENTITY_SCORE is 5 or more. As you can see from the picture, there are nearly 30000 reports for just those three months.
You can inspect the SYMPTOM_TEXT in the table, but you can also click on the URL in the first column to see the full information on the medalerts website.
Click here to see the CSV file
Compare this with the Incredible Hulk report, and you can see quite clearly that the reports with such entity scores are very descriptive, quite technical and very likely reported by a medical professional.
How long does it take to run the script?
A reader asked me how long it takes to run the script. Since the Python code just loops through the entire dataframe row by row, and spaCy loads the SYMPTOM_TEXT into a document for each row, it can be a bit slow.
So you should only do this type of analysis offline and store the results.
Here are some numbers for how much time it took to run the script on my MacBook laptop:
August 2022: 7500 rows = 236 seconds
July 2022: 13200 rows = 400 seconds
June 2022: 13200 rows = 467 seconds
May 2022: 19000 rows = 679 seconds
I cannot say for sure this is a linear relationship in terms of number of rows in the dataframe, but it looks quite close. :-)
What about the weekly VAERS update?
Since VAERS is updated each week, I would compare the SYMPTOM_TEXT for the latest snapshot with the SYMPTOM_TEXT for the current row (which you are using to display the ENTITY_SCORE) and only process the document if the text has changed.
That is, you would put this line of code within an if block which tests for the condition that SYMPTOM_TEXT_NEW != SYMPTOM_TEXT_OLD
doc_med = nlp_medical(symptom_text)This will help you avoid recomputing the score for the same identical document for each update.